Sparse/Dense QUBO Matrix in OpenJij#
The number of nonzero elements in the interaction matrix of the Ising Hamiltonian or QUBO matrix can vary greatly depending on the problem.
Many real-life problems have sparse interaction matrices with relatively few nonzero values.
Therefore, by default, OpenJij internally keeps the interaction coefficients as a sparse matrix.
However, depending on the problem, many elements of the interaction matrix may be non-zero.
In this case, data retention in sparse matrix form will slow down the computation.
To solve this problem, OpenJij provides a parameter, sparse, that allows users to control how the matrix is retained.
In this tutorial, we will explain how to use sparse. We will also compare the speed of keeping data in a sparse matrix with that of keeping data in a dense matrix.
import openjij as oj
import numpy as np
import time
The internal retention of data can be configured as follows:
sampler = oj.SASampler()
response = sampler.sample_ising(h, J, sparse = True)
With sparse = True, the sampler retains the interaction matrix as a sparse matrix.
The latest OpenJij has sparse = True by default (as of 2022/10 (v0.6.5)).
You can change this flag by:
response = sampler.sample_ising(h, J, sparse = False)
This would make internal data retention a dense matrix.
Comparing Execution Speed in Sparse and Dense Mode#
Next, let us examine how data retention can affect execution time.
NUM_READS = 1
BETA_MAX = 14000000
BETA_MIN = 0.0015
num_spins = [100,200,500,1000,2000,5000]
p_list = [0.1,0.3,0.5,0.8]
A variant of the Sherrington-Kirkpatrick (SK) model without external fields is used here as a benchmark. The SK model without external fields is a model with the following Hamiltonian:
where is a random Gaussian noise with a mean of 0 and variance of 1. In the SK model, all elements follow a random distribution. However, here we will examine how computation time changes as we vary the fraction of elements in the interaction matrix and make the interaction sparse.
First, we set up a random interaction. Here, the parameters are the number of spins and the fraction of elements in the interaction matrix .
def create_random_interaction(n,p):
h,J = {},{}
for i in range(n-1):
for j in range(i+1, n):
if np.random.random() <= p:
# OpenJij keeps data in the form of an upper triangular matrix.
J[i, j] = np.random.normal(0, 1) / np.sqrt(n)
return J,h
We first calculate the case with a dense matrix. Here, we change the number of spins , and the proportion of elements in the interaction matrix is also varied .
# Benchmark OpenJij Dense
print('OpenJij Dense')
sampler = oj.SASampler()
openjij_dense_time = []
openjij_dense_energy = []
for p in p_list:
for n in num_spins:
J,h = create_random_interaction(n,p)
start = time.perf_counter()
response = sampler.sample_ising(h, J, num_sweeps=1000, num_reads=NUM_READS, beta_max=BETA_MAX, beta_min=BETA_MIN, sparse = False)
elapsed_time = time.perf_counter() - start
openjij_dense_time.append(elapsed_time)
openjij_dense_energy.append(np.mean(response.energies))
print(f"p = {p}, n = {n} : \telapsed_time:{elapsed_time}[sec]")
OpenJij Dense
p = 0.1, n = 100 : elapsed_time:0.008780483999998978[sec]
p = 0.1, n = 200 : elapsed_time:0.02667508199999702[sec]
p = 0.1, n = 500 : elapsed_time:0.08442160699996748[sec]
p = 0.1, n = 1000 : elapsed_time:0.2422231150000016[sec]
p = 0.1, n = 2000 : elapsed_time:0.8939402400000063[sec]
p = 0.1, n = 5000 : elapsed_time:5.163792424999997[sec]
p = 0.3, n = 100 : elapsed_time:0.007489820999978747[sec]
p = 0.3, n = 200 : elapsed_time:0.023477394999986245[sec]
p = 0.3, n = 500 : elapsed_time:0.09813072299999703[sec]
p = 0.3, n = 1000 : elapsed_time:0.29128573799999913[sec]
p = 0.3, n = 2000 : elapsed_time:1.025661466000031[sec]
---------------------------------------------------------------------------
KeyboardInterrupt Traceback (most recent call last)
Cell In[4], line 9
7 for p in p_list:
8 for n in num_spins:
----> 9 J,h = create_random_interaction(n,p)
10 start = time.perf_counter()
11 response = sampler.sample_ising(h, J, num_sweeps=1000, num_reads=NUM_READS, beta_max=BETA_MAX, beta_min=BETA_MIN, sparse = False)
Cell In[3], line 5, in create_random_interaction(n, p)
3 for i in range(n-1):
4 for j in range(i+1, n):
----> 5 if np.random.random() <= p:
6 # OpenJij keeps data in the form of an upper triangular matrix.
7 J[i, j] = np.random.normal(0, 1) / np.sqrt(n)
9 return J,h
KeyboardInterrupt:
Next, we examine the case with a sparse matrix. The execution conditions are the same as for the dense matrix case.
# Benchmark OpenJij Sparse
print('OpenJij Sparse')
sampler = oj.SASampler()
openjij_sparse_time = []
openjij_sparse_energy = []
for p in p_list:
for n in num_spins:
J,h = create_random_interaction(n,p)
start = time.perf_counter()
response = sampler.sample_ising(h, J, num_sweeps=1000, num_reads=NUM_READS, beta_max=BETA_MAX, beta_min=BETA_MIN, sparse=True)
elapsed_time = time.perf_counter() - start
openjij_sparse_time.append(elapsed_time)
openjij_sparse_energy.append(np.mean(response.energies))
print(f"p = {p}, n = {n} : \telapsed_time:{elapsed_time}[sec]")
OpenJij Sparse
p = 0.1, n = 100 : elapsed_time:0.015455876999993734[sec]
p = 0.1, n = 200 : elapsed_time:0.01172593000001143[sec]
p = 0.1, n = 500 : elapsed_time:0.05383983299998363[sec]
p = 0.1, n = 1000 : elapsed_time:0.18197211800000446[sec]
p = 0.1, n = 2000 : elapsed_time:0.727910971[sec]
p = 0.1, n = 5000 : elapsed_time:4.896766718999999[sec]
p = 0.3, n = 100 : elapsed_time:0.007563688999994156[sec]
p = 0.3, n = 200 : elapsed_time:0.023680912000003218[sec]
p = 0.3, n = 500 : elapsed_time:0.12488942999999608[sec]
p = 0.3, n = 1000 : elapsed_time:0.663864641999993[sec]
p = 0.3, n = 2000 : elapsed_time:2.272948852999974[sec]
p = 0.3, n = 5000 : elapsed_time:14.29901092199998[sec]
p = 0.5, n = 100 : elapsed_time:0.012703374000011536[sec]
p = 0.5, n = 200 : elapsed_time:0.052227919000017664[sec]
p = 0.5, n = 500 : elapsed_time:0.19213491300001806[sec]
p = 0.5, n = 1000 : elapsed_time:0.8431962399999975[sec]
p = 0.5, n = 2000 : elapsed_time:3.5385885899999607[sec]
p = 0.5, n = 5000 : elapsed_time:23.98366295200003[sec]
p = 0.8, n = 100 : elapsed_time:0.01682115800002748[sec]
p = 0.8, n = 200 : elapsed_time:0.07765323400002444[sec]
p = 0.8, n = 500 : elapsed_time:0.5316407230000095[sec]
p = 0.8, n = 1000 : elapsed_time:1.358698779000008[sec]
p = 0.8, n = 2000 : elapsed_time:5.622073602[sec]
p = 0.8, n = 5000 : elapsed_time:37.54809538400002[sec]
Let us plot the results.
import matplotlib.pyplot as plt
fig = plt.figure(figsize=(10,6))
sparse_time = np.array(openjij_sparse_time).reshape(len(p_list),-1)
dense_time = np.array(openjij_dense_time).reshape(len(p_list),-1)
color_list = ["blue", "red", "green", "orange"]
for index,p in enumerate(p_list):
plt.plot(num_spins,dense_time[index], '-o', label=f'dense $p$ ={p}', color = color_list[index], markersize=8)
plt.plot(num_spins,sparse_time[index], '-^', label=f'sparse $p$ ={p}', color = color_list[index], markersize=8)
plt.xscale("log")
plt.yscale("log")
plt.xlabel('number of spins', fontsize=18)
plt.ylabel('time [sec]', fontsize=18)
plt.legend(fontsize=12, ncol = 4, bbox_to_anchor=(0.7, 0.7, 0.3, 0.5))
plt.show()
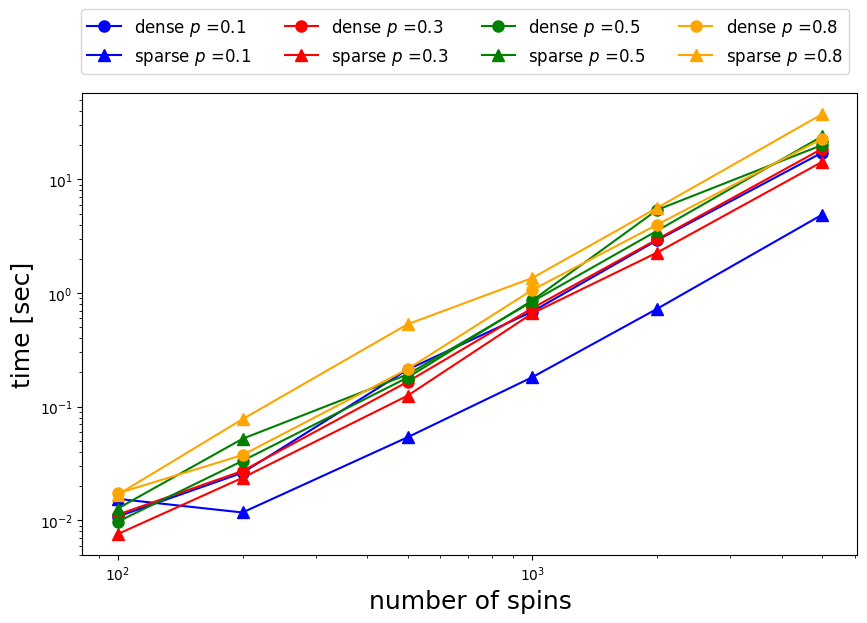
When the fraction of elements packed in the interaction matrix is , the sparse matrix case with sparse=True is several times faster than the dense matrix case.
However, at around , both speeds are approximately the same.
Furthermore, the dense matrix is faster when .
Thus, we can see that execution speed varies depending on whether a sparse or dense matrix is used.
As a result, you will achieve faster results by switching accordingly to the problems.